
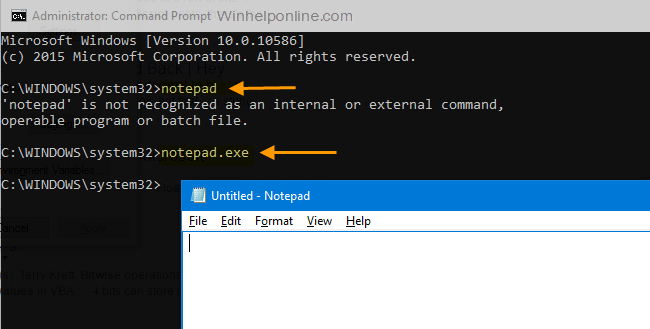
- #How to run a program in command prompt windows 10 software#
- #How to run a program in command prompt windows 10 series#
- #How to run a program in command prompt windows 10 simulator#
MS-GF+ (aka MSGF+ or MSGFPlus) performs peptide identification by scoring MS/MS spectra against peptides derived from a protein sequence database. from Sequest, XTandem, Inspect, or MSGF+) and merges that information with the corresponding MASIC results files, appending the relevant MASIC stats for each peptide hit result. Reads the contents of a tab-delimited peptide hit results file (e.g. MASIC (MS/MS Automated Selected Ion Chromatogram generator) Generates selected ion chromatograms (SICs) for all of the parent ions chosen for fragmentation in an LC-MS/MS analysis. Features are annotated by glycan family relationships and in-source fragmentation patterns. Features are characterized by monoisotopic mass, elution time, and isotopic fit score.

#How to run a program in command prompt windows 10 software#
The software uses a list of glycan targets to search for expected features in MS1 spectra. GlyQ-IQ is software that performs a targeted, chromatographic centric search of mass spectral data for glycans.
#How to run a program in command prompt windows 10 series#
Dat file and parse out the information for each entry, creating a series of tab delimited text files or creating a FASTA file.įormularity is software for assignment of low weight molecular formula from high-resolution mass spectra. The Uniprot DAT File Parser can read a Uniprot. The Protein Sequence Motif Extractor reads a fasta file or tab delimited file containing protein sequences, then looks for the specified motif in each protein sequence.
#How to run a program in command prompt windows 10 simulator#
The Protein Digestion Simulator can be used to read a text file containing protein or peptide sequences ( FASTA format or delimited text) then output the data to a tab-delimited file. Although the splitting is random, each section will have a nearly identical number of residues.Ī Population Variation plug-in for the Skyline software program that can assist researchers in determining whether their target peptides have known mutations in the general human population. Software Category: Fasta File, Protein Sequence, or Protein Database Related toolsĬonsole application that reads a protein FASTA file and splits it apart into a number of sections. The Visual Integration for Bayesian Evaluation (VIBE) software is a visualization tool that allows the user to observe classification accuracies at the class level and evaluate classification accuracies on any subset of available data types based on the posterior probability models defined for the individual and integrated data. VIBE: Visual Integration for Bayesian Evaluation The Protein Coverage Summarizer can be used to determine the percent of the residues in each protein sequence that have been identified.Ī high-performance multiprocessor implementation of the NCBI BLAST library.ĭraws correctly proportioned and positioned two and three circle Venn diagrams (aka Euler diagrams) whose colors can be customized and the diagrams copied to the clipboard or saved to disk. The ListPOR program (List Parser for Outlier Removal) can be used to read a file containing columns of grouped values and remove outlier values using Grubb’s test. Windows graphical user interface tool for viewing LC-MS data and identifications. Software Category: Data Analysis and Data Presentation ToolsĪctive Data Canvas is a web-based visual analytic tool to visualize data matrix (expression matrix) and for users to interactively identify the structured domain knowledges (e.g., pathways and other genesets) linked to a cluster.Īctive Data Canvas was hosted at, but that was turned off due to lack of use. VIPER (Visual Inspection of Peak/Elution Relationships) can be used to visualize and characterize the features detected during LC-MS analyses. InfernoRDN can perform various downstream data analysis, data reduction, and data comparison tasks including normalization, hypothesis testing, clustering, and heatmap generation.Īligns multiple LC-MS datasets to one another after which LC-MS features can be matched to a database of peptides (typically an AMT tag database) This information is used to subtract out errors from parent ion protonated masses.

Reduces mass measurement errors for parent ions of tandem MS/MS data by modeling systematic errors based on putative peptide identifications. Used to de-isotope mass spectra and to detect features from mass spectrometry data using observed isotopic signatures. All of our open source software is posted at our group’s GitHub Repository.


 0 kommentar(er)
0 kommentar(er)
